We've just release our version of PICRUSt for the rumen microbiome. This software uses the 16S sequences from the Global Rumen Census and the nearly 500 published genomes from cultured rumen organisms currently available (most from the Hungate 1000 project) to allow functional inferences from 16S meta-taxonomic studies and can be found at www.cowpi.org.
The paper has now been published in Frontiers in Microbiology here as part of a special research topic on "Metaomic Approaches to Study the Rumen Microbiome: Challenges and Innovation". We demonstrate how using the data directly from the Rumen allows a better prediction of the abundance of functional units (for example see figure 1 from the paper, reproduced here).
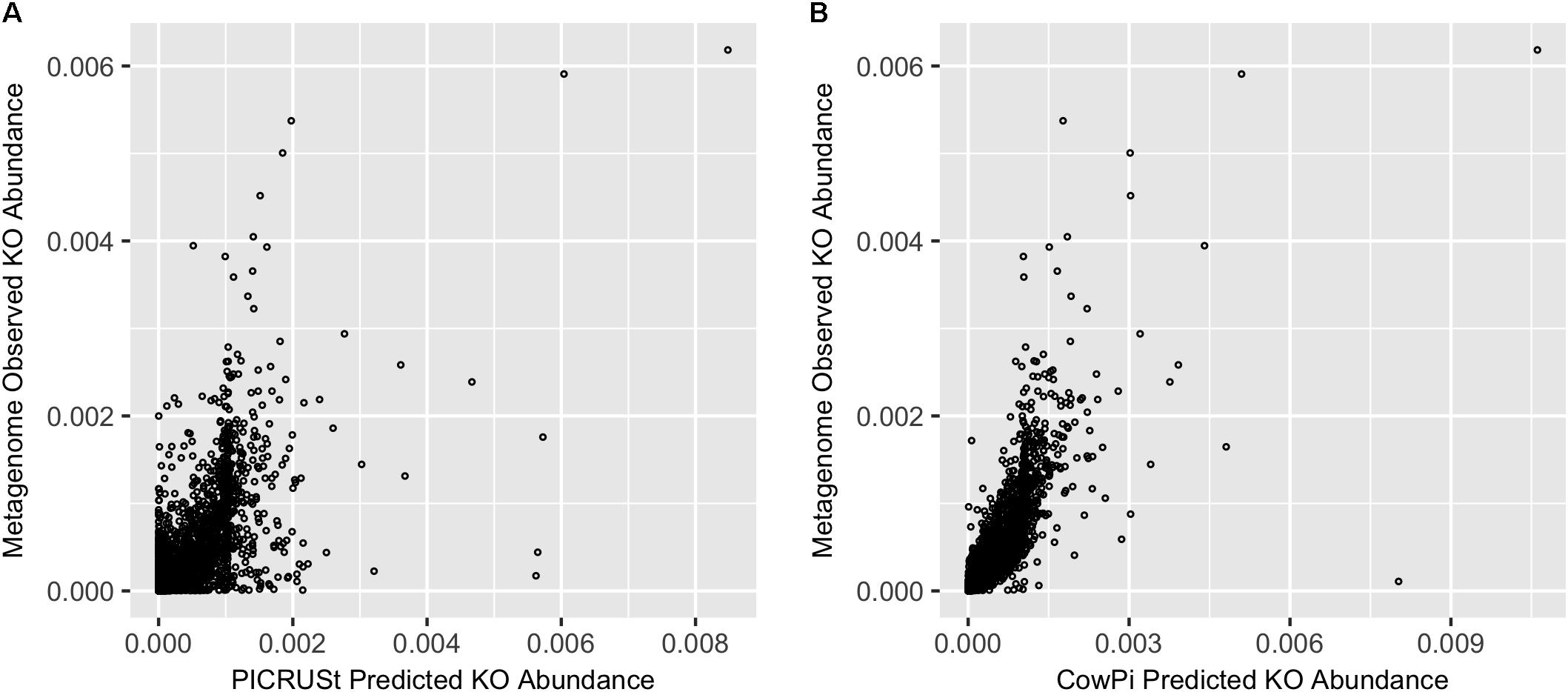 |
| FIGURE 1. Prediction accuracy on the Hess et al. (2011) dataset. (A) Metagenome compared with PICRUSt and (B) Metagenome compared with CowPI. Points represent the relative abundance of KOs in the observed (y-axis) and predicted (x-axis) dataset |
The data behind the tool can be downloaded at Zenodo under this DOI:
All users of this system will need to register and set up their account (walk through tutorial here). Once set up the impatient users can assess a rapid overview of using the system here and a more detailed explanation of each step of the workflow can be found here.
We would welcome feedback from the community and would encourage other groups to implement external-facing version for other groups to use so please get in touch if you have any questions/suggestions.
